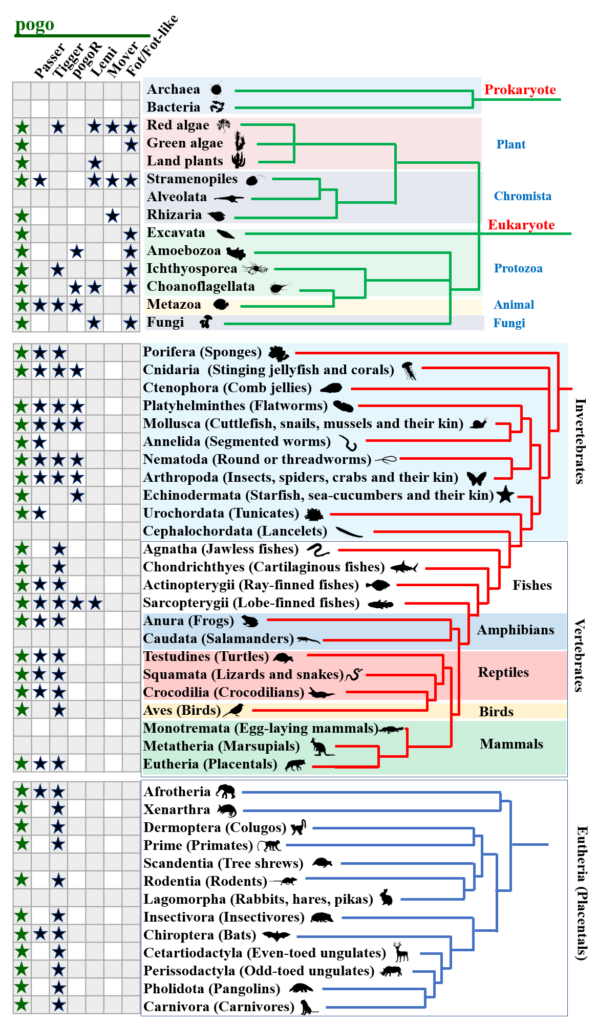
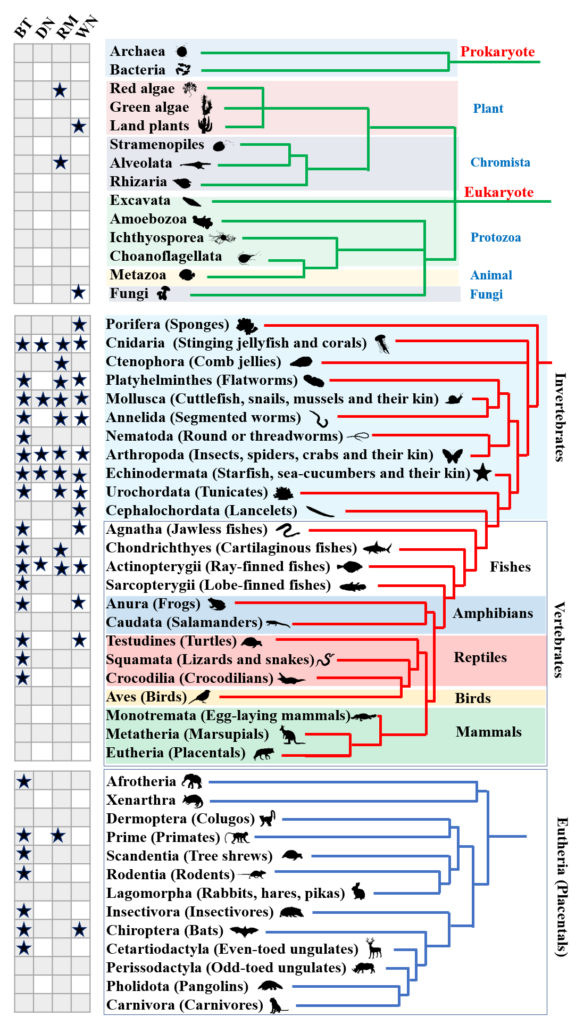
Editing Efficiency Prediction
CRISPR与TnpB双系统的设计平台CRISPy-web
TnpB Editing Efficiency Predictor (TEEP): https://go.tnpb.app/
Protein Mutational Effect Predictor (ProMEP): https://github.com/wenjiegroup/ProMEP
EVOLVEpro: https://github.com/mat10d/EvolvePro
Bridge RNA Design Tool: https://arcinstitute.org/tools/bridge
CRISPR
CRISPR Integrated DNA Technologies | IDT
Chopchop: http://chopchop.cbu.uib.no/
CCTOP: CCTop – CRISPR/Cas9 target online predictor (uni-heidelberg.de)
CRISPR-CAS++ (paris-saclay.fr)
CRISPRloci – CRISPR-Cas annotation (uni-freiburg.de)
crisprSQL – CRISPR off-target database
CRISPR RGEN Tools (rgenome.net)
https://benchling.com/pub/c2c2
http://crispr-analyzer.dkfz.de/
http://www.e-crisp.org/E-CRISP/
http://crispresso.pinellolab.org/submission
http://guides.sanjanalab.org/#/
https://www.crisprindelphi.design/
Addgene: CRISPR Plasmids and Resources
TALEN
Tools | TAL Effector Nucleotide Targeter 2.0 (cornell.edu)
SAPTA TAL Targeter | Tools | tDnA (rice.edu)
ZNF
Zinc Finger Tools (scripps.edu)
The Zinc Finger Consortium | Software Tools (zincfingers.org)