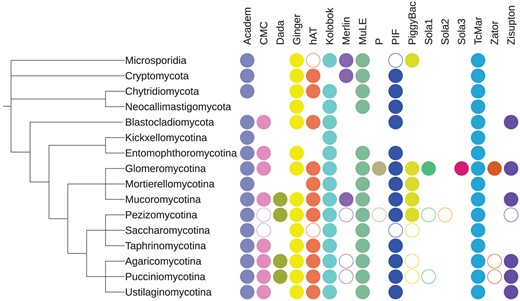
Distribution of TEs across the eukaryote phylogeny

Structure and taxonomy of eukaryotic TEs

Reference: Wells JN, Feschotte C. A Field Guide to Eukaryotic Transposable Elements. Annu Rev Genet. 2020 Nov 23;54:539-561. doi: 10.1146/annurev-genet-040620-022145.
Taxonomic distribution of the 17 DDE Tn superfamilies across the eukaryotic tree of life.

Yuan YW, Wessler SR. The catalytic domain of all eukaryotic cut-and-paste transposase superfamilies. Proc Natl Acad Sci U S A. 2011 May 10;108(19):7884-9. doi: 10.1073/pnas.1104208108. Epub 2011 Apr 25. PMID: 21518873; PMCID: PMC3093488.
The assignment of protein domains to DNA TE superfamilies in Fungi
| Superfamily | RepBase (Fungi) | Count in RepBase | Yuan and Wessler | DNA TE (with domain) | Observed Distribution | Domains |
|---|---|---|---|---|---|---|
| Academ | Only Puccinia graminis | 7 | n | 9,709 | Low copy, in most taxa, highest abundance in Pucciniomycotina | RNase H-like (Yuan and Wessler 2011) |
| CMC | Basidiomycota and Mucorales | 28 | y | 11,961 | Broader distribution, expanded in Agaricomycetes Pucciniomycotina and Mucoromycotina | Transposase_21: PF02992, Transposase_23: PF03017*, Transposase_24: PF03004* |
| Dada | Only Laccaria bicolor | 2 | — | 1,023 | Broader distribution (4 phyla) | RNase H-like (Kojima and Jurka 2013) |
| Ginger | Only Malassezia globosa | 1 | n | 6,648 | Ubiquitous, expansions in Dikarya | rve: PF00665 |
| hAT | Only Dikarya | 37 | y | 33,376 | Ubiquitous | Dimer_Tnp_hAT: PF05699*, DUF659: PF04937, DUF4371: PF14291, DUF4413: PF14372* |
| KDZ (Zisupton) | Only Puccinia graminis | 4 | — | 14,607 | Basidiomycota, Rhizophagus, Mucoromycotina, and Allomyces | RNase H-like (Iyer et al. 2014) |
| Kolobok | Only Rhizophagus irregularis | 5 | n | 3,214 | Low copy, ubiquitous, highest abundance in R. irregularis | RNase H-like (Yuan and Wessler 2011) |
| Merlin | Only Rhizopus oryzae | 5 | y | 4,255 | Single occurrences in Dikarya, expansions in Microsporidia | DDE_Tnp_IS1595: PF12762 |
| MULE | Dikarya and Rhizopus | 36 | y | 17,658 | Ubiquitous | Transposase_mut: PF00872, MULE: PF10551 |
| Novosib | n | 0 | n | 0 | Only copies without transposase | — |
| P | Only Pucciniales & Allomyces | 17 | y | 11 | Single occurrences | Tnp_P_element_C: PF12596*, Tnp_P_element: PF12017 |
| PIF/Harb | Diverse Fungi | 76 | y | 13,443 | Ubiquitous | Plant_tran: PF04827, DDE_Tnp_4: PF13359 |
| PiggyBac | Mucor and Pezizomycotina | 4 | y | 5,965 | Mucoromycota, Microsporidia, and Pezizomycotina | DDE_Tnp_1_7: PF13843 |
| Sola1 | n | 0 | n | 140 | Rhizophagus, single occurrences in Dikarya | RNase H-like (Majorek et al. 2014) |
| Sola2 | n | 0 | n | 1 | One case in Aspergillus flavus | RNase H-like (Majorek et al. 2014) |
| Sola3 | n | 0 | n | 637 | Only Rhizophagus | RNase H-like (Majorek et al. 2014) |
| Tc1/Mariner | Diverse Fungi | 148 | y | 93,120 | Ubiquitous | DDE_1: PF03184, DDE_3: PF13358, Transposase_1: PF01359 |
| Transib | n | 0 | n | 0 | Absent | RAG1: PF12940 |
| Zator | Only Puccinia striiformis | 2 | n | 1,165 | Rhizophagus, single occurrences in Basidiomycota | RNase H-like (Majorek et al. 2014) |
Taxonomic distribution of DNA TE superfamilies in major fungal lineages