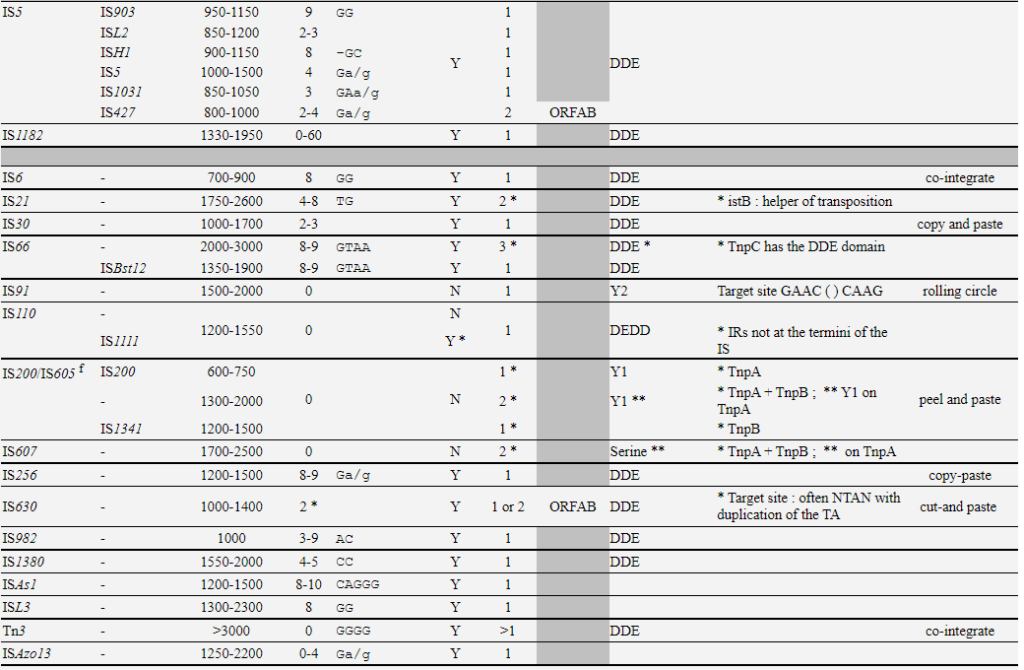
Copy-out–Paste-in IS elements identifed in prokaryotes
| Families | groups | Size-range | DR | Ends | IRs | Nb ORF | Frameshift | Chemistry |
|---|---|---|---|---|---|---|---|---|
| IS1 | IS1 | 740-1180 | 8-9 | GGnnnTG | Y | 2 | ORFAB | DDE |
| ISMhu11 | 900-4600 | 0-10 | Y | 2 | ORFAB | |||
| IS3 | IS911 | 1250 | Y | 2 | ORFAB | |||
| IS150 | 1200-1600 | 3-4 | TG | Y | 2 | ORFAB | DDE | |
| IS407 | 1100-1400 | 4 | TG | |||||
| IS51 | 1000-1400 | 3-4 | TG | |||||
| IS3 | 1150-1750 | 3-4 | TGa/g | |||||
| IS2 | 1300-1400 | 5 | TG | |||||
| IS481 | – | 950-1300 | 4-15 | TGT | Y | 1 | ||
| IS30 | – | 1000-1700 | 2-3 | Y | 1 | DDE | ||
| IS110 | IS110 | 1200-1550 | 0 | N | DEDD | |||
| IS1111 | Y * | DEDD | ||||||
| IS256 | – | 1200-1500 | 8-9 | Ga/g | Y | 1 | DDE | |
| ISL3 | – | 1300-2300 | 8 | GG | Y | 1 | ||
| IS21 | – | 1750-2600 | 4-8 | TG | Y | 2 * | DDE | |
| ISLre2 |
Classification, terminal features, TSD features and the number of entries of DNA transposons in Repbase
| Group | Superfamily | Termini | Mechanism | TSD | Entries |
|---|---|---|---|---|---|
| IS630/Mariner | Mariner/Tc1 | YR..YR | Cut-and-paste | TA | 2,539 |
| Zator | GG..CC | Cut-and-paste | 3 | 54 | |
| IS481/Ginger | Ginger1 | TGT..ACA | Cut-and-paste | 4 | 39 |
| Ginger2/TDD | TGT..ACA | Cut-and-paste | 4–5 | 20 | |
| IS3/IS3EU | IS3EU | TAY..RTA | Copy-out–Paste-in? | 6 | 23 |
| IS1016/Merlin | Merlin | GG..CC | Cut-and-paste | 8–9 | 75 |
| IS256/DxxH | hAT | YA..TR | Cut-and-paste | 5–8 | 2,955 |
| MuDR | GR..YC | Cut-and-paste | 8–9 | 1,345 | |
| P | CA..TG | Cut-and-paste | 7–8 | 189 | |
| Kolobok | RR..YY | Cut-and-paste | TTAA | 286 | |
| Dada | ? | 6–7 | 36 | ||
| IS1380/piggyBac | piggyBac | YY..RR | Cut-and-paste | TTAA | 377 |
| IS5/PHIS | Harbinger | RR..YY | Cut-and-paste | 3 | 1,097 |
| ISL2EU | RR..YY | Cut-and-paste? | 2 | 88 | |
| Spy | Cut-and-paste | no | |||
| NuwaI | |||||
| NuwaII | |||||
| Pangu | |||||
| CCHH | EnSpm/CACTA | CAC..GTG | Cut-and-paste | 2–4 | 715 |
| Transib | CAC..GTG | Cut-and-paste | 5 | 123 | |
| KDZP | Zisupton | ? | ? | 8 | 18 |
| Sola | Sola | Cut-and-paste | |||
| Sola1 | ? | Cut-and-paste | 4 | 100 | |
| Sola2 | GRG..CYC | Cut-and-paste | 4 | 90 | |
| Sola3 | GAG..CTC | Cut-and-paste | TTAA | 28 | |
| Unclassified Sola | Cut-and-paste | 1 | |||
| ? | Academ | YR..YR | Cut-and-paste | 3–4 | 90 |
| ? | Novosib | CA..TG | Cut-and-paste | 8 | 9 |
| Crypton | Crypton | Copy-out–Paste-in | |||
| CryptonF | Copy-out–Paste-in | 0 | 23 | ||
| CryptonA | TTA.. | Copy-out–Paste-in | 0 | 17 | |
| CryptonI | ? | Copy-out–Paste-in | 0 | 9 | |
| CryptonS | TATGG.. | Copy-out–Paste-in | 0 | 59 | |
| CryptonV | ? | Copy-out–Paste-in | 0 | 46 | |
| Unclassified Crypton | Copy-out–Paste-in | 80 | |||
| Helitron | Helitron | TC..CTRR | Rolling circle | 0 | 955 |
| Polinton | Polinton | AG..CT | Self-synthesizing | 6 | 108 |
| Unclassified DNA transposon | 2,357 | ||||
| Total | 13,960 |
Eukaryotic cut-and-paste transposase superfamilies
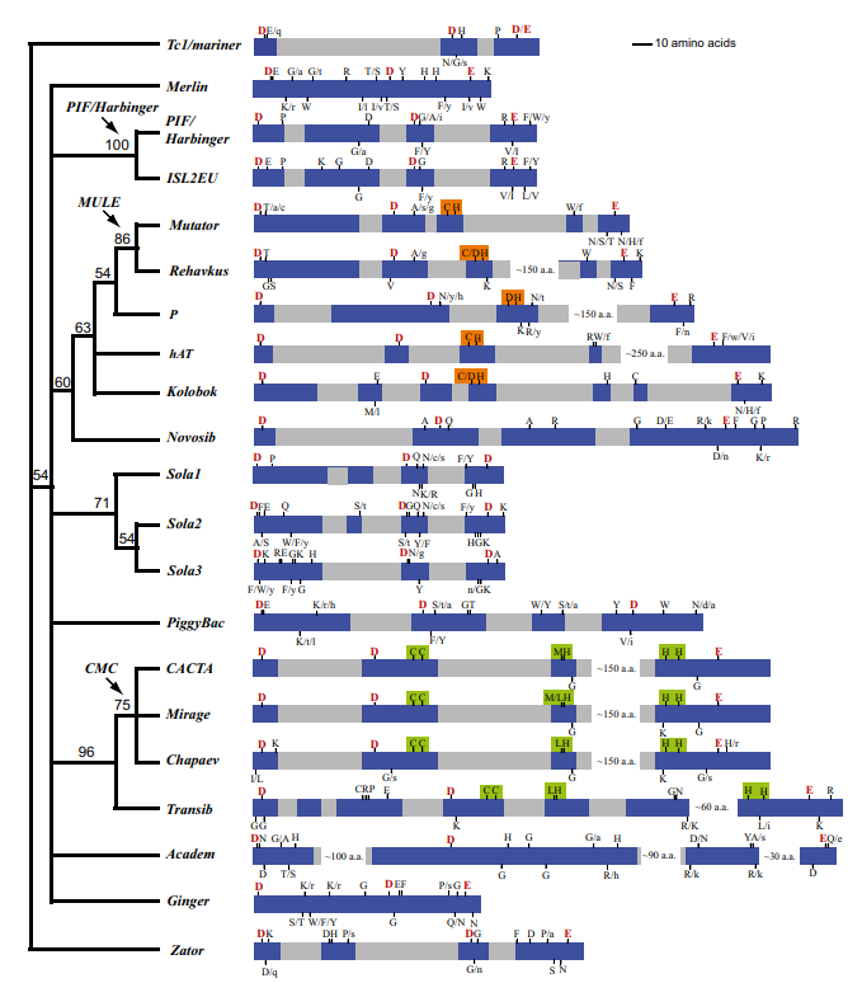
Fig. 2. An unrooted consensus tree of the transposase superfamilies inferred from the presence or absence of the highly conserved residues in the signature strings. Bootstrap values are at the nodes. The arrows with labels indicate superfamily clusters merged in our revised classification. Shown on the right is a schematic representation of the DDE/D domain and the signature string for each superfamily. Conserved blocks are highlighted in blue, variable regions are in gray. White gaps are regions not drawn to scale. The DDE triads are highlighted in red. Alternative residues are marked by slashes; lowercase indicates that a residue occurs in <10% of the sequences in the alignment profile. The C/DH motif is highlighted in orange; the C(2)C, [M/L]H, and H(3-4)H motifs are highlighted in green.
List of Copy-out–Paste-in IS elements
List of mobile elements whose transposases have been examined by secondary structure prediction programs
| Family | Element (or protein) analyzed | Active or # copies in genome1 | From secondary structure, type of DDE/D motif2 | Relevant references3 |
| IS1 | IS1NISSto9 | >40*5 | DD(24)EDD(20)E | * Nyman et al., 1981; Ohta et al., 2002, 2004; Siguier et al., 2009 |
| IS1595 | 1. ISPna2 | ?,DD(36)N” | Siguier et al., 2009 | |
| 2. ISH4 | ?,DD(36)E” | Siguier et al., 2009 | ||
| 3. IS1016C | ?,DD(34)E” | Siguier et al., 2009 | ||
| 4. IS1595 | ?,DD(35)N” | Siguier et al., 2009 | ||
| 5. ISSod11 | 13 | DD(34)H | Siguier et al., 2009 | |
| 6. ISNWi1 | ?,DD(35)E” | Siguier et al., 2009 | ||
| 7. ISNha5 | ?,DD(33)E” | Siguier et al., 2009 | ||
| Merlin: MERLIN1_SM | Consensus | DD(36)E | Feschotte, 2004 | |
| IS3 | IS911 | Active | DD(35)E | Polard and Chandler, 1995; Rousseau et al., 2002 |
| IS481 | IS481 | ?00* | DD(35)E | *Glare et al., 1990; Chandler and Mahillon, 2002 |
| IS4 | IS50R | Active | PDB ID: 1muhDD(-strand)E | Rezshazy et al., 1993; Davies et al., 2000 |
| IS701 | IS701ISRso17 | Active (15*)7 | DD(-strand)E | *Mazel et al., 1991 |
| ISH3 | ISC1359ISC1439A | 513 | DD(-strand)E | |
| IS1634 | IS1634ISMac5ISPlu4 | Active (?0*)77 | DD(-strand)E | *Vilei et al., 1999 |
| IS5 | IS903 | Active | DD(65)E | Derbyshire et al., 1987; Rezshazy et al., 1993; Tavakoli et al., 1997 |
| PIF/Harbinger: PIFa (Z. mays) | Active | DD(59)E | Zhang et al., 2001; Kapitonov and Jurka, 2004; Sinzelle et al., 2008 | |
| IS1182 | IS660ISPsy6 | 314 | DD(-strand)E | Takami et al., 2001 |
| IS6 | IS6100 | Active | DD(34)E | Martin et al., 1990; Mahillon and Chandler, 1998 |
| IS21 | IS21 | Active | DD(45)E | Mahillon and Chandler, 1998; Berger and Haas, 2001 |
| IS30 | IS30 | Active | DD(33)E | Caspers et al., 1984; Mahillon and Chandler, 1998 |
| IS66 | IS679ISPsy5ISMac8 | Active333 | DD(-helical?)E | Han et al., 2001 |
| IS110 | IS492IS1111 | Active20 | DEDDDEDD | Perkins-Balding et al., 1999; Buchner et al., 2005 |
| IS256 | IS256 | Active | DD(-helical)E | Mahillon and Chandler, 1998; Prudhomme et al., 2002 |
| MuDr/Foldback (Mutator) | Active | DD(-helical)E | Eisen et al., 1994; Babu et al., 2006; Hua-Van and Capy, 2008 | |
| IS630 | ISY100 | Active | DD(34)E | Doak et al., 1994; Feng and Colloms, 2007 |
| Tc1/mariner: Mos1 (D. mauritiana) | Active | PDB ID: 2f7tDD(34)D | Plasterk et al., 1999; Richardson et al., 2006 | |
| Zator: Zator-1_HM | 36* | DD(43)E | *Bao et al., 2009 | |
| IS982 | ISPfu3 | 5 | DD(47)E | Mahillon and Chandler, 1998 |
| IS1380 | IS1380A | ?00* | DD(-strand)E | *Takemura et al., 1991; Chandler and Mahillon, 2002 |
| piggyBac (T. ni) | Active | DD(-strand)D | Cary et al., 1989; Sarkar et al., 2003; Mitra et al., 2008 | |
| ISAs1 | ISAzo3 | 7 | DD(-strand)E/D? | |
| ISL3 | IS31831IS651 | Active22 | DD(-helical)E | Suzuki et al., 2006 |
| Tn3 | Tn3 (E. coli) | Active | DD(-helical?)E | Grindley, 2002 |
| hAT | Hermes (M. domestica) | Active | PDB ID: 2bw3 DD(-helical)E insertion | Warren et al., 1994; Rubin et al., 2001; Hickman et al., 2005 |
| CACTA | CACTA1 (A. thaliana) En/Spm ZM | Active | DD(-helical?)E/D? | Miura et al., 2001; DeMarco et al., 2006 |
| P | Drosophila | Active | ? | Rio, 2002 |
| Transib | Transib1_AG | Consensus | DD(-helical)E | Kapitonov and Jurka, 2005; Chen and Li, 2008 |
| RAG1 (M. musculus) | Active | DD(-helical)E | Kim et al., 1999; Landree et al., 1999; Lu et al., 2006 | |
| Sola | Sola3-3_HM | Multiple copies* | DD(40)E | *Bao et al., 2009 |
Hickman AB, Chandler M, Dyda F. Integrating prokaryotes and eukaryotes: DNA transposases in light of structure. Crit Rev Biochem Mol Biol. 2010 Feb;45(1):50-69. doi: 10.3109/10409230903505596.
Classification, distribution and the number of entries of LTR retrotransposons in Repbase
| Superfamily | Total |
|---|---|
| Copia | 10,595 |
| Gypsy | 6,694 |
| BEL | 1,855 |
| ERV | |
| ERV1 | 1,967 |
| ERV2 | 1,266 |
| ERV3 | 657 |
| ERV4 | 187 |
| Lentivirus | 4 |
| Unclassified ERV | 325 |
| Unclassified LTR | 719 |
| DIRS | 418 |
Classification, and the number of entries of non-LTR retrotransposons in Repbase
| Group | Clade | Total |
|---|---|---|
| CRE | CRE | 43 |
| R2 | R4 | 46 |
| Hero | 23 | |
| NeSL | 106 | |
| R2 | 159 | |
| Dualen | RandI/Dualen | 13 |
| L1 | Proto1 | 6 |
| L1 | 1,690 | |
| Tx1 | 273 | |
| RTE | RTETP | 1 |
| Proto2 | 47 | |
| RTEX | 138 | |
| RTE | 487 | |
| I | Outcast | 23 |
| Ingi | 17 | |
| Vingi | 141 | |
| I | 195 | |
| Nimb | 108 | |
| Tad1 | 141 | |
| Loa | 74 | |
| R1 | 237 | |
| Jockey | 243 | |
| CR1 | Rex1 | 95 |
| CR1 | 803 | |
| Kiri | 91 | |
| L2 | 285 | |
| L2A | 5 | |
| L2B | 27 | |
| Crack | 140 | |
| Daphne | 227 | |
| Ambal | Ambal | 8 |
| Penelope | Penelope | 477 |
| SINE | SINE1/7SL | 95 |
| SINE2/tRNA | 539 | |
| SINE3/5S | 30 | |
| SINEU | 17 | |
| Unclassified SINE | 112 | |
| Unclassified non-LTR retrotransposon | 179 | |
| Total | 7,341 |
Reference: Kenji K. Kojima, Structural and sequence diversity of eukaryotic transposable elements, Genes & Genetic Systems, 2019, Volume 94, Issue 6, Pages 233-252
Proposal of TE classes with some members having a DNA transposon phenotype

Reference: Benoît Piégu, Solenne Bire, Peter Arensburger, Yves Bigot, A survey of transposable element classification systems – A call for a fundamental update to meet the challenge of their diversity and complexity,
Molecular Phylogenetics and Evolution, Volume 86, 2015, Pages 90-109,
ISSN 1055-7903, https://doi.org/10.1016/j.ympev.2015.03.009.
Major Features of Prokaryotes IS families (ISfinder)

Overview of common transposon annotation tools
| Approach | Class I | Class II | ||||||||
| Name | Novo. | Struc. | Simil. | LTR | LINE | SINE | TIR | HEL | MITE | |
| RepeatMasker | x | x | x | x | x | x | x | x | ||
| RepeatModeler | x | x | x | x | x | x | x | |||
| CLARI_TE | (107) | x | x | x | x | x | x | x | x | x |
| TESeeker | (41) | x | x | x | x | x | x | x | ||
| PILER | (40) | x | x | x | x | x | x | x | ||
| Censor | (108) | x | x | x | x | x | x | x | ||
| RepLong | (109) | x | x | x | x | x | x | x | ||
| EDTA | (44) | x | x | x | x | x | x | x | x | x |
| MGEScan | (110) | x | x | x | x | x | x | |||
| LTR_Finder | (111) | x | x | |||||||
| LtrDetector | (112) | x | x | |||||||
| LTRpred | (73) | x | x | x | x | |||||
| LTRharvest | (66) | x | x | x | x | |||||
| LTRdigest | (113) | x | x | |||||||
| SINE-Finder | (68) | x | x | x | ||||||
| SINE-Scan | (69) | x | x | x | ||||||
| TIRvish | (67) | x | x | |||||||
| HelitronScanner | (42) | x | x | |||||||
| MUSTv2 | (70) | x | x | |||||||
| MiteFinderII | (71) | x | x | |||||||
| MITE-Tracker | (72) | x | x | |||||||
| detectMITE | (45) | x | x | |||||||
| MITE-Hunter | (47) | x | x | |||||||
| TransposonUltimate | ||||||||||
